3D Voronoï tessellations applied to protein structures
Voro3D is an original tool which provides a brand new point of view on protein 3D structures. In order to perform a 3D Voronoï tessellation, Voro3D uses as input a protein structure file in the PDB format. After calculation, different structural properties of interest like secondary structures assignment, environment accessibility, contact matrices can be derived without any geometrical cut-off. Voro3D provides also a visualization of these tessellations superimposed on the associated protein structure, from which it is possible to model a polygonal protein surface or to quantify the contact areas between a protein and a ligand, for instance.
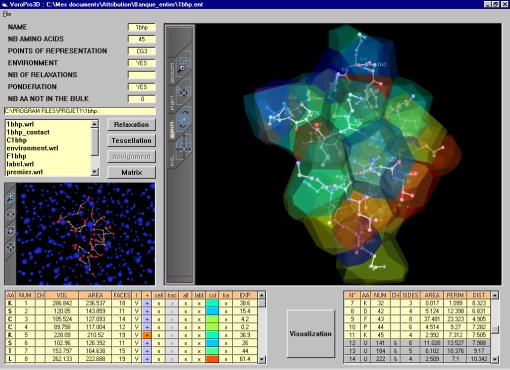
An example of Voro3D screen :
Starting from any PDB file, Voro3D allows to :
More information ? See these articles :
- Select the points representing amino acids (alpha carbons, geometric centers)
- Embed your protein structure in a model environment made of a relaxed random packing of spheres
Choose the sphere radius as well as the number of environment layers
Relax this environment as many times as you want
- Perform the Voronoi tessellation or the Laguerre polyhedral decomposition (weighted Voronoi tessellation). In the latter case it is possible to choose different weights
- Visualize the structure and its environment
- Visualize the tessellation superimposed on the protein structure, modify several graphic parameters
- Derive all the values concerning the cells and the cell faces
- Assign automatically the secondary structure elements according to the VoTAP procedure
Derive environment accessibility and contact matrices- Export the visualization files (*.wrl) to internet or to your PowerPoint presentations thanks to the VRML viewer
To install this programm on your PC (windows 98, 2000, NT, XP) :
- Soyer, A., Chomilier, J., Mornon J.P., Jullien, R. and Sadoc, J.F. (2000). Voronoi tessellation reveals the condensed matter character of folded proteins. Phys Rev Lett, 85, 3532-3535.
- Angelov, B., Sadoc, J.F., Jullien, R., Soyer, A., Mornon, J.P. and Chomilier, J. (2002). Nonatomic solvent-driven Voronoi tessellation of proteins : an open tool to analyse protein folds. Proteins, 49, 446-456.
- Sadoc, J.F., Jullien, R. and Rivier, N. (2003). The Laguerre polyhedral decomposition : application to protein folds. Eur. Phys. J. B., 33, 355-363.
- Dupuis, F., Sadoc, J.F. and Mornon, J.P. (2004). Protein secondary structure assignment through Voronoi tessellation. Proteins, 55, 519-528
Voro3D was designed by Franck Dupuis in collaboration with :
- download the file voro3Dsetup.zip (or here)
- uncompress it
- double click on Setup.exe
- then double click on cortvrml.exe
- Jean-Paul Mornon, Laboratoire de Minéralogie Cristallographie Paris, Equipe de Biologie Structurale, Université Paris 6 et 7, France
- Jean-François Sadoc, Laboratoire de Physique des Solides, Université Paris 11, Orsay, France
- Rémi Jullien, Laboratoire des Verres, Université de Montpellier 2, Montpellier, France
- Borislav Angelov Institute of Biophysics, Bulgarian Academy of Sciences, Acad. G. Bonchev Str., Block 21, Sofia 1113, Bulgaria